




2026 2025 2024 2023 2022 2021 2017-2020 2010-2016
2026
JC. Bean, J. Jian, TC. Lu, H. Liu, KM. McDermott, DA. Threat, SV. Jossy, ME. Burt, J. Cheng, Y. Deng, X. Fang, X. Geng, J. Han, Y. Li, H. Liu, Q. Liu, Y. Liu, Y. Shi, L. Tu, M. Wang, X. Xu, Y. Yang, M. Yu, X. Liu, M. Sun, F. Wang, OZ. Ginnard, Y. Yang, Y. He, C. Wang, Y. Qi, H. Li# (co-corresponding), Y. Xu# Sex-specific differences in arcuate nucleus of the hypothalamus in response to nutritional states. (2026) Nature Communications [link]
GA. Perez, Z. Lai, GA. Edwards III, JM. Dundee, SN. Leahy, C. Qi, Y. Qi, YJ. Park, TC. Lu, M. Uddin, R. Zhao, H. Zheng, H. Li, JL. Jankowsky# Neuronal subtype governs amyloid structure, cellular response, and cognitive outcome in genetically targeted APP mouse models. (2026) Molecular Neurodegeneration [link][PDF]Submitted/preprint:
BB. Su*, T. Jackson,* W. Blackmon*, H. Ames, C. Holt, A. Anagnostou, V. Szafron, S. Anvari, H. Li# (co-corresponding), CM. Davis# Gamma Delta T Cells mediate shrimp allergy through regulatory cytokine pathways. submitted
H. Liu*#, Q. Liu*, JC. Bean*, F. Wang, Y. Li, KM. Conde, M. Wang, Y. Deng, Y. Yang, T. Zhou, J. Cheng, JJ. Jian, Y. Liu, M. Sun, X. Liu, J. Qiu, X. Wu, L. Xue, Q. Tong, BR. Arenkiel, M. Xue, H. Cui, TC. Lu, Y. Qi, H. Li, C. Wang, Y. He, Y. Yang, L. Tu, Y. Xu# Lef1+ hypothalamic neurons mediate cold responses via Kcnk2. submitted
AMJ. Adams, KLK. Wu, Z. Li, P. Li, S. Alimukhamedov, A. Nguyen, H. Pan, Z. Huang, S. Migliarini, JF. Haberberger, R. Dear, J. Zhu, L. Huang, B. Sun, ZCK. Chan, J. Liang, H. Li, R. Gong, P. Vertes, M. Pasqualetti, R. Lin, A. Isakova#, J. Ren# Developmental transcriptomic trajectories define forebrain-projecting serotonin system organization. submitted
S. Gibson, S. Deal, Y. Park, B. Sun, Y. Qi, J. Mok, H. Chung, H. Li, S. Yamamoto# Hippo signaling regulates cuticle pigmentation and dopamine metabolism in Drosophila. submitted [bioRxiv]
O. Pena-Ramos, M. Gedam, X. Zhang, S. Wang, C. Qi, T. Huang, C. Rajpurohit, M. Dunlap, Y. Deng, N. Auld, S.Y. Jung, H. Li, J. Li, L. Luo, J. Peng, C. Cheng, H. Zheng# Astrocyte cell-surface proteomics identified CD44 as an OPN/SPP1 receptor regulating lipid metabolism and glial crosstalk in Alzheimer’s disease. submitted
X. Li, N. Zang, Q. Zhao, Y. Sun, U. S., M. Hua, W. Zhou, Y. Qi, H. Chen, L. Xia, J. Qin, L. Sun, C. Cheng, Z. Liu, L. Chen, Y. Xu, H. Li, X. Hou, Y. He, X. Zhuang, Z. Sun# High-level estrogen impairs memory through ER via a hypothalamic-hippocampal neural circuit. submitted
D. Park, RM. Lawrence, T. Jackson, H. Li, JC. Mills# Evidence that injury can cause Drosophila gut differentiated, polyploid enterocytes to be recruited as stem cells via paligenosis. submitted
Y. Furuta, Hu Chen, J. Jia, TC. Lu; J. Wang, Y. Qi, M. Maletic-Savatic, H. Li, Z Liu, J. Chin# Single-nucleus transcriptomics reveals mechanisms underlying epileptiform activity-induced dysregulation of neurogenesis in early Alzheimer’s disease. submitted
2025
YJ. Park*, TC. Lu*, T. Jackson, LD. Goodman, L. Ran, J. Chen, CY. Liang, E. Harrison, C. Ko, X. Chen, B. Wang, AL. Hsu, E. Ochoa, K. F. Bieniek, S. Yamamoto, Y. Zhu, H. Zheng, Y. Qi, HJ. Bellen#, H. Li# (co-corresponding) Distinct systemic impacts of Aβ42 and Tau revealed by whole-organism snRNA-seq. (2025) Neuron [PDF][PDF+Supplement][link][AD-FCA website][CBS news1][news2][news3][news4][news5][news6]
OV. Goldman#, AE. DeFoe, Y. Qi, Y. Jiao, S-C. Weng, L. Houri-Zeevi, P. Lakhiani, T. Morita, J. Razzauti, A. Rosas-Villegas, YN. Tsitohay, MM. Walker, BR. Hopkins, Mosquito Cell Atlas Consortium, OS. Akbari, LB. Duvall, H. White-Cooper, TR. Sorrells, R. Sharma, H. Li# (co-corresponding), LB. Vosshall#, N. Shai# Mosquito Cell Atlas: A single-nucleus transcriptomic atlas of the adult Aedes aegypti mosquito. (2025) Cell [PDF][link][data portal]
JB. Chen*, MC. Wang*, S. Gong*, H. Li# (corresponding) Toward precision longevity: aging interventions in the single-cell atlas era. (2025) Frontiers in Aging (review)[PDF][link]
M.P. Marques, B. Sun, YJ. Park, T. Jackson, TC. Lu, Y. Qi, E. Harrison, M.C. Wang, K. Venkatachalam, O.M. Pasha, A. Varanasi, D.K. Carey, D.R. Mani, J. Zirin, M. Qadiri, Y. Hu, N. Perrimon, S.A. Carr, N.D. Udeshi, L. Luo, J. Li, H. Li# (corresponding) In-situ glial cell-surface proteomics identifies pro-longevity factors in Drosophila. (2025) eLife [link]
Y. Liu*, H. Liu*, HX. Wong, N. Auld, KM. Conde, Y. Li, M. Yu, Y. Deng, Q. Liu, X. Fang, M. Wang, Y. Shi, OZ. Ginnard, Y. Yang, L. Tu, H. Liu, JC. Bean, J. Han, ME. Burt, SV. Jossy, Y. Yang, C. Wang, Q. Tong, BR. Arenkiel, H. Li, Y. Xu#, Y. He# Prolonged exposure to food odors suppresses feeding via an olfactory bulb-to-hypothalamus circuit. (2025) Nature Communications [link][PDF]
S. Wang, C. Qi, C. Rajpurohit, B. Ghosh, W. Xiong, B. Wang, Y. Qi, SH. Hwang, BD. Hammock, H. Li, L. Gan, H. Zheng# Inhibition of Soluble Epoxide Hydrolase Confers Neuroprotection and Restores Microglial Homeostasis in a Tauopathy Mouse Model.(2025) Molecular Neurodegeneration [link][PDF]
M. Qadiri*, Y. Liu*, AR. Kim, M. Han, E. Zhou, A. Veal, TC Lu, H. Li, Y. Hu#, N. Perrimon# FlyPhoneDB2: A computational framework for analyzing cell-cell communication in Drosophila scRNA-seq data integrating AlphaFoldmultimer predictions.(2025) Computational and Structural Biotechnology Journal [link][PDF]
C. Lyu*, Z. Li*, C. Xu, KKL. Wong, DJ. Luginbuhl, CN. McLaughlin, Q. Xie, T. Li, H. Li, L. Luo# Dimensionality reduction simplifies synaptic partner matching in an olfactory circuit.(2025) Science [link][PDF]
2024
S Gao, Y Qi, Q Zhang, A Mohammed, Y Lee, Y Guan, H Li# (co-corresponding), Y Fu#, M Wang# Aging Atlas Reveals Cell-Type-Specific Regulation of Pro-longevity Strategies. (2024) Nature Aging [link][PDF][webportal link]
B Wang*, H Martini-Stoica*, C Qi*, TC Lu, S Wang, W Xiong, Y Qi, Y Xu, MSardiello, H Li, H Zheng# TFEB–vacuolar ATPase signaling regulates lysosomal function and microglial activation in tauopathy. (2024) Nature Neuroscience [link] [PDF]
DC Sutton, JC Andrews, DM Dolezal, YJ Park,H Li, DF Eberl, S Yamamoto#, AK Groves# Comparative exploration of mammalian deafness gene homologues in the Drosophila auditory organ shows genetic correlation between insect and vertebrate hearing (2024) PLOS One [link][PDF]
C Xu, Z Li, C Lyu, Y Hu, CN McLaughlin, KK Wong, Q Xie, DJ Luginbuhl, H Li, ND Udeshi, T Svinkina, DR Mani, S Han, T Li, Y Li, R Guajardo, AY Ting, SA Carr, J Li#, L Luo# Molecular and Cellular Mechanisms of Teneurin Signaling in Synaptic Partner Matching.(2024) Cell [link] [PDF]
2023
TC Lu*, M Brbić*, YJ Park, T Jackson, J Chen, SS Kolluru, Y Qi, NS Katheder, XT Cai, S Lee, YC Chen, N Auld, CY Liang, S Ding, D Welsch, S D’Souza, AO Pisco, RC Jones, J Leskovec, EC Lai, HJ Bellen, L Luo, H Jasper#, SR Quake#, H Li# (co-corresponding) Aging Fly Cell Atlas Identifies Exhaustive Aging Features at Cellular Resolution. (2023) Science [link][PDF][Supplement PDF]
[AFCA website][news]
A Mamiya, A Sustar, I Siwanowicz, Y Qi, TC Lu, P Gurung, C Chen, JS Phelps, AT Kuan, A Pacureanu, WA Lee, H Li, N Mhatre, JC Tuthill# Origins of proprioceptor feature selectivity and topographic maps in the Drosophila leg. (2023) Neuron [link] [PDF]
A Raz*, G Vida*, S Stern*, S Mahadevaraju*, J Fingerhut*, J Viveiros*, S Pal*, J Grey*, M Grace*, C Berry, H Li, J Janssens, W Saelens, Z Shao, C Hun, Y Yamashita, T Przytycka, B Oliver, J Brill, H Krause, E Matunis, H White-Cooper, S DiNardo#, M Fuller# Emergent dynamics of adult stem cell lineages from single-nucleus and single-cell RNA-Seq of Drosophila testes. (2023) eLife [link][PDF]
H Liu, H Li, Y Xu Hunger extends lifespan by modulating histone proteins. (2023) Life Metabolism[link][PDF]
NS. Katheder, KC. Browder, D Chang, AD Mazière, P Kujala, S Dijk, J Klumperman, TC Lu, H Li, Z Lai, D Sangaraju, H Jasper# Nicotinic acetylcholine receptor signaling maintains epithelial barrier integrity. (2023) eLife [link][PDF]
2022
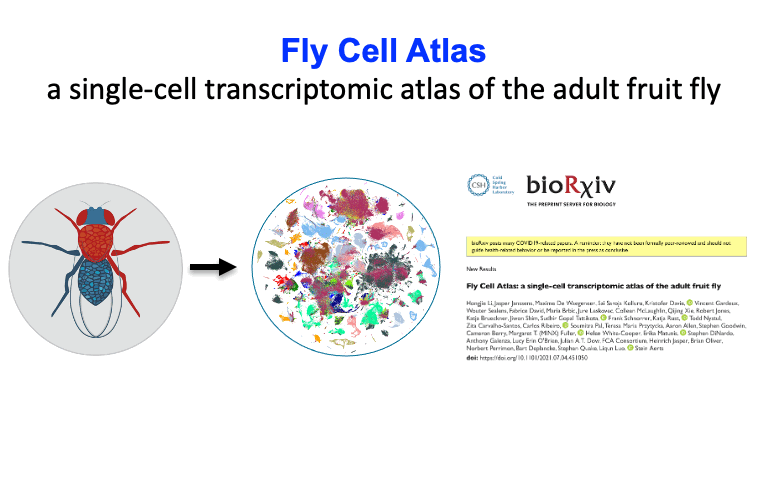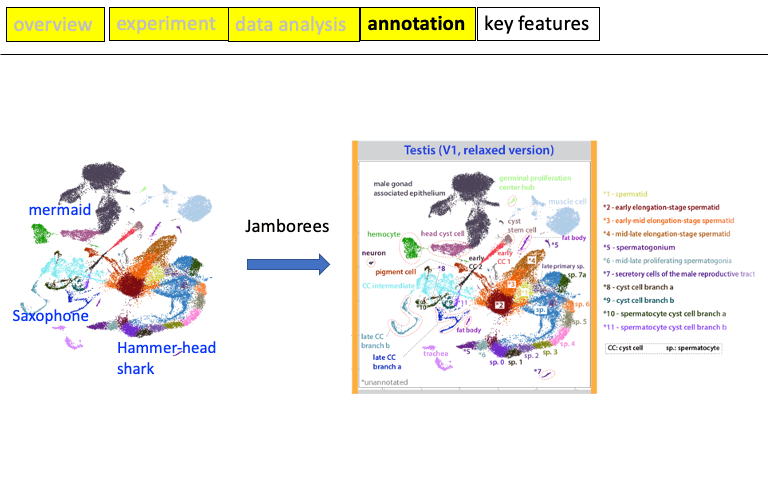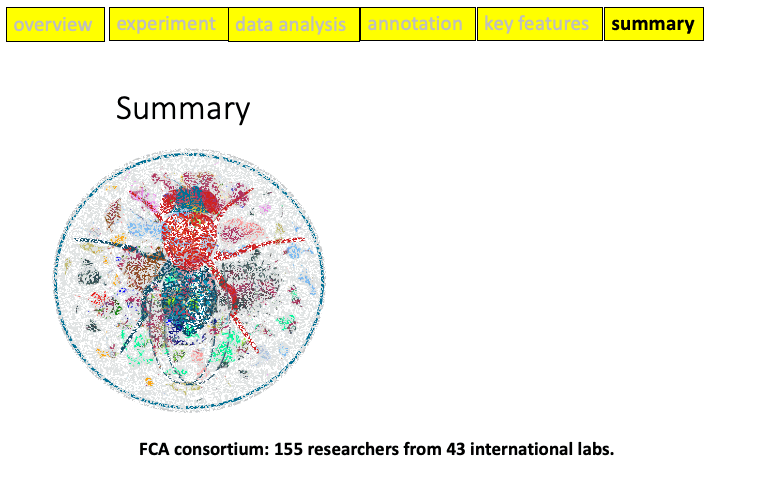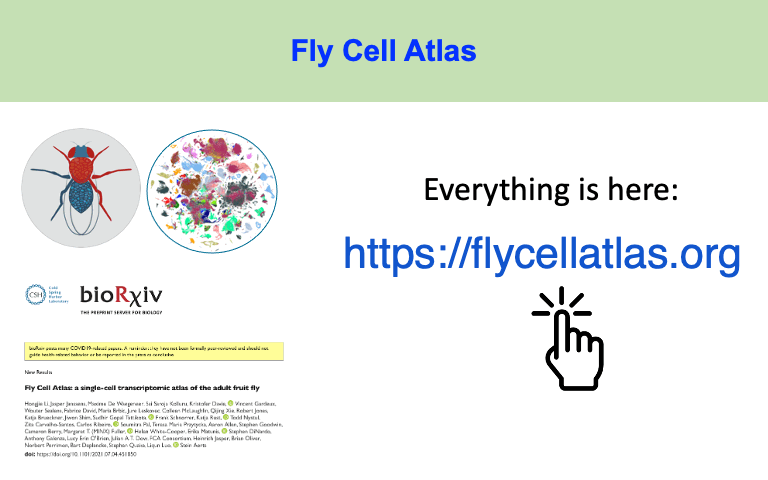
H Li*, J Janssens*, M Waegeneer, S Kolluru, K Davie, V Gardeux, W Saelens, F David, M Brbić, K Spanier, J Leskovec, C McLaughlin, Q Xie, R Jones, K Brueckner, J Shim, S Tattikota, F Schnorrer, K Rust, T Nystul, Z Carvalho-Santos, C Ribeiro, S Pal, S Mahadevaraju, T Przytycka, A Allen, S Goodwin, C Berry, M Fuller, H White-Cooper, E Matunis, S DiNardo, A Galenza, L O’Brien, J Dow, D Agarwal, Y Ahmed-Braimah, M Arbeitman, MM Ariss, J Augsburger, K Ayush, CC Baker, T Banisch, K Birker, R Bodmer, B Bolival, SE Brantley, JA Brill, NC Brown, NA Buehner, XT Cai, R Cardoso-Figueiredo, F Casares, A Chang, TR Clandinin, S Crasta, C Desplan, AM Detweiler, DB Dhakan, E Donà, S Engert, S Floc’hlay, N George, AJ González-Segarra, AK Groves, S Gumbin, Y Guo, DE Harris, Y Heifetz, SL Holtz, F Horns, B Hudry, RJ Hung, YN Jan, JS Jaszczak, GSXE Jefferis, J Karkanias, TL Karr, NS Katheder, J Kezos, AA Kim, SK Kim, L Kockel, N Konstantinides, TB Kornberg, HM Krause, AT Labott, M Laturney, R Lehmann, S Leinwand, J Li, JSS Li, K Li, K Li, L Li, T Li, M Litovchenko, HH Liu, Y Liu, TC Lu, J Manning, A Mase, M Matera-Vatnick, NR Matias, CE McDonough-Goldstein, A McGeever, AD McLachlan, P Moreno-Roman, N Neff, N Neville, S Ngo, T Nielsen, CE O’Brien, D Osumi-Sutherland, MN Özel, I Papatheodorou, M Petkovic, C Pilgrim, AO Pisco, C Reisenman, EN Sanders, G Dos Santos, K Scott, A Sherlekar, P Shiu, D Sims, RV Sit, M Slaidina, HE Smith, G Sterne, YH Su, D Sutton, M Tamayo, M Tan, I Tastekin, C Treiber, D Vacek, G Vogler, S Waddell, W Wang, RI Wilson, MF Wolfner, YE Wong, A Xie, J Xu, S Yamamoto, J Yan, Z Yao, K Yoda, R Zhu, RP Zinzen, H Jasper, B Oliver, N Perrimon#, B Deplancke#, S Quake#, L Luo#, S Aerts# Fly Cell Atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. (2022) Science [link] [PDF][Supplement PDF]
[FlyCellAtlaswebsite]
C McLaughlin*, Y Qi*, S Quake#, L Luo#, H Li# (co-corresponding) Isolation and RNA sequencing of single nuclei from Drosophila tissues. (2022) STAR Protocols [link][PDF]
D Task, C Lin, A Vulpe, A Afify, S Ballou, M Brbić, P Schlegel, J Raji, G. Jefferis, H Li, K Menuz, C Potter# Chemoreceptor Co-Expression in Drosophila Olfactory Neurons. (2022) eLife [link][PDF]
Q Xie*, J Li*, H Li, ND Udeshi, T Svinkina, S Kohani, R Guajardo, DR Mani, C Xu, T Li, S Han, W Wei, SA Shuster, DJ Luginbuhl, AY Ting, SA Carr, L Luo# Transcription factor control of dendrite targeting via combinatorial cell-surface codes. (2022) Neuron[link][PDF]
J Xu*, Y Liu*, H Li, A Tarashansky, C Kalicki, R Hung, Y Hu, A Comjean, S Kolluru, B Wang, S Quake, L Luo, A McMahon, J A.T. Dow, N Perrimon# Transcriptional and functional motifs defining renal function revealed by single nuclear sequencing. (2022) PNAS [link][PDF]
M Herre*, O Goldman*, TC Lu, G Caballero-Vidal, Y Qi, Z Gilbert, Z Gong, T Morita, S Rahiel, M Ghaninia, R Ignell, B Matthews, H Li, L Vosshall, M Younger*# Non-Canonical Odor Coding in the Mosquito. (2022) Cell [link][PDF]
S Lee, Y Chen, FCA Consortium, A Gillen, J Taliaferro, B Deplancke, H Li, EC Lai# Diverse cell-specific patterns of alternative polyadenylation in Drosophila. (2022) Nature Communications [link][PDF]
2021
McLaughlin*, C., Brbic*, M., Xie, Q., Li, T., Horns, F., Kolluru, S.S., Kebschull, J., Vacek, D., Xie, A., Li, J., Jones R., Leskovec, J., Quake S.#, Luo, L.#,Li, H. Single-cell transcriptomes of developing and adult olfactory receptor neurons in Drosophila. (2021) eLife [link] [PDF]
Xie, Q., Brbic, M., Horns, F., Kolluru, S.S., Jones R., Li, J., Reddy, A., Xie, A., Kohani S., Li, Z., McLaughlin, C., Li, T., Xu, C., Vacek, D., Luginbuhl, D., Leskovec, J., Quake S.#, Luo, L.#,Li, H. Temporal evolution of single-cell transcriptomes of Drosophila olfactory projection neurons. (2021) eLife [link][PDF]
Li, T.#, Fu, TM., Li, H., Xie, Q., Luginbuhl, D., Betzig, E., Luo, L# Cellular Bases of Olfactory Circuit Assembly Revealed by Systematic Time-Lapse Imaging.(2021) Cell [link] [PDF]
Cai, X., Li, H., Borch Jensen, M., Maksoud, E., Borneo, J., Liang, Y., Quake S., Luo, L., Pejmun, P., and Jasper, H# Gut cytokines modulate olfaction through metabolic reprogramming of glia. (2021) Nature [link] [PDF]
2017-2020
Li, H.*, Horns, F.*, Wu, B., Xie, Q., Li, J., Li, T., Luginbuhl, D.J., Quake, S.R.#, and Luo, L.# Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. (2017) Cell [link] [PDF]
Li, H.,*#(co-corresponding), Li, T*., Horns, F., Li, J., Xie, Q., Xu, C., Wu, B., Kebschull, J.M., McLaughlin, C.N., Kolluru, S.S., Jones R.J., Vacek, D., Xie, A., Luginbuhl, D.J., Quake S.R.#, Luo, L.# Single-cell transcriptomes reveal diverse regulatory strategies for olfactory receptor expression and axon targeting (2020) Current Biology [link] [PDF]
Li, H.*, Shuster, S.A.*, Li, J.*, and Luo, L.# Linking neuronal lineage and wiring specificity. (2018) Neural Development. (Review) [link] [PDF]
Li, H.# (corresponding) Single-cell RNA sequencing in Drosophila: Technologies and applications. (2020) Wiley Development Biology (invited Review) [link] [PDF]
Li, J.*#, Han, S.*, Li, H., Udeshi, N.D., Svinkina, T., Mani, D.R., Xu, C., Guajardo, R., Xie, Q., Li, T., Luginbuhl, D.J., Wu, B., McLaughlin, C.N., Xie, A., Kaewsapsak, P., Quake S.R., Carr, S.A., Ting, A.Y.#, Luo, L.# Cell-Surface Proteomic Profiling in the Fly Brain Uncovers Wiring Regulators. (2020) Cell [link] [PDF]
Wenderski, W., Wang, L., Krokhotin, A., Walsh, J.J., Li, H., Shoji, H., Ghosh, S., George, R.D., Miller, E.L., Elias, L., Gillespie, M.A., Son, E.Y., Staahl, B.T., Baek, S.T., Stanley, V., Moncada, C., Shipony, Z., Linker, S.B., Marchetto, M.C, Gage, F.G., Chen, D., Sultan, T., Zaki, M.S., Ranish, J.A., Miyakawa, T., Luo, L., Malenka R.C., Crabtree, G.R.#, Gleeson, J.G.# Loss of the neural-specific BAF subunit ACTL6B relieves repression of early response genes and causes recessive autism. (2020) PNAS [link] [PDF]
Tracy Cai, X., Li, H., Safyan, A., Gawlik, J., Pyrowolakis, G., and Jasper, H.# AWD regulates timed activation of BMP signaling in intestinal stem cells to maintain tissue homeostasis. (2019) Nature Communications [link] [PDF]
Xie, Q.*, Wu, B.*, Li, J., Xu, C., Li, H., Luginbuhl, D.J., Wang, X., Ward, A., and Luo, L.# Transsynaptic Fish-lips signaling prevents misconnections between nonsynaptic partner olfactory neurons. (2019) PNAS [link] [PDF]
Li, J., Guajardo, R., Xu, C., Wu, B., Li, H., Li, T., Luginbuhl, D.J., Xie, X., and Luo, L.# Stepwise wiring of the Drosophila olfactory map requires specific Plexin B levels. (2018) eLife [link] [PDF]
2010-2016
Li, H., Qi, Y., and Jasper, H.# Preventing Age-Related Decline of Gut Compartmentalization Limits Microbiota Dysbiosis and Extends Lifespan. (2016) Cell Host & Microbe [link] [PDF]
Li, H., Qi, Y., and Jasper, H.# Ubx dynamically regulates Dpp signaling by repressing Dad expression during copper cell regeneration in the adult Drosophila midgut. (2016) Development Biology [link] [PDF]
Li, H., and Jasper, H.# Gastrointestinal stem cells in health and disease: from flies to humans.(2016) Disease Models & Mechanisms (Review) [link] [PDF]
Li, H.,# (co-corresponding) and Wei, C.# Diet, gut microbiota and obesity. (2015)J Nutrition Health Food Sci (Review) [link] [PDF]
Li, H., Qi, Y., and Jasper, H.# Dpp signaling determines regional stem cell identity in the regenerating adult Drosophila gastrointestinal tract. (2013) Cell Reports [link] [PDF]
Ayyaz, A., Li, H., and Jasper, H.# Haemocytes control stem cell activity in the Drosophila intestine. (2015) Nature Cell Biology [link] [PDF]
Hine, C.M., Li, H., Xie, L., Mao, Z., Seluanov, A.#, and Gorbunova, V.# Regulation of Rad51 promoter. (2014) Cell Cycle [link] [PDF]
*(co-)first, #(co-)corresponding
Fly cell atlas